Reducing code runtime is important for developers. Python Profilers, like cProfile helps to find which part of the program or code takes more time to run. This article will walk you through the process of using cProfile module for extracting profiling data, using the pstats module to report it and snakeviz for visualization.
By the end of this post, you will know:
- Why do we need Python Profilers ?
- Introduction to cProfile
- How to use cProfile ?
- Profiling a function that calls other functions
- How to use Profile class of cProfile
- How to export cProfile data ?
- How to visualize cProfile reports?
- Profiling Linear Regression Model from scikit learn
1. Why do we need Python Profilers ?
Today, there are so many of areas where you write code ranging from basic conditional logics to complex websites, apps, algorithms, etc. The main aspect while writing any code, especially when deploying, is that it should consume the lowest computational time and cost.
This is especially important when you run code on cloud services like AWS, Google Cloud or Azure, where there is a defined cost associated with the usage of computing resources. If you have two pieces of code that give the same result, the one that takes the least time and resource is usually chosen.
Let’s say you have an algorithm that takes a lot of time to run. And you want to reduce the code run time. The first question that might crop up is:
Why does my code take so long to run?
Python Profilers can answer that question. It tells you which part of the code took how long to run. This lets you focus on that particular part and achieve efficiency. I cover in detail how to use Python profiler, particularly ‘cProfile’, with various examples.
2. Introduction to cProfile
cProfile is a built-in python module that can perform profiling. It is the most commonly used profiler currently.
But, why cProfile is preferred?
- It gives you the total run time taken by the entire code.
- It also shows the time taken by each individual step. This allows you to compare and find which parts need optimization
- cProfile module also tells the number of times certain functions are being called.
- The data inferred can be exported easily using
pstatsmodule. - The data can be visualized nicely using
snakevizmodule. Examples come later in this post.
That’s a lot of useful information. Let’s look at the code example to use cProfile. Start by importing the package.
# import module
import cProfile
3. How to use cProfile ?
cProfile provides a simple run() function which is sufficient for most cases. The syntax is cProfile.run(statement, filename=None, sort=-1).
You can pass python code or a function name that you want to profile as a string to the statement argument.
If you want to save the output in a file, it can be passed to the filename argument. The sort argument can be used to specify how the output has to be printed. By default, it is set to -1( no value).
Let’s call cProfile.run() on a simple operation.
import numpy as np
cProfile.run("20+10")
Output:
3 function calls in 0.000 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.000 0.000 <string>:1(<module>)
1 0.000 0.000 0.000 0.000 {built-in method builtins.exec}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
Above you passed a simple addition code as a statement to the run() function of cProfile. Let’s understand the output.
Line no.1: shows the number of function calls and the time it took to run.
Line no.2: Ordered by: standard name means that the text string in the far right column was used to sort the output. This could be changed by the sort parameter.
Line no. 3 onwards contain the functions and sub functions called internally. Let’s see what each column in the table means.
ncalls: Shows the number of calls madetottime: Total time taken by the given function. Note that the time made in calls to sub-functions are excluded.percall: Total time / No of calls. ( remainder is left out )cumtime: Unliketottime, this includes time spent in this and all subfunctions that the higher-level function calls. It is most useful and is accurate for recursive functions.- The
percallfollowingcumtimeis calculated as the quotient ofcumtimedivided by primitive calls. The primitive calls include all the calls that were not included through recursion.
You could see that it isn’t very complex because the operation we did is simple.
4. Profiling a function that calls other functions
Now let’s try profiling on a code that calls other functions. In this case, you can pass the call to main() function as a string to cProfile.run() function.
# Code containing multiple dunctions
def create_array():
arr=[]
for i in range(0,400000):
arr.append(i)
def print_statement():
print('Array created successfully')
def main():
create_array()
print_statement()
if __name__ == '__main__':
cProfile.run('main()')
Output:
Array created successfully
400041 function calls in 0.091 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.004 0.004 0.091 0.091 <ipython-input-10-4dd6137cfe06>:12(main)
1 0.059 0.059 0.087 0.087 <ipython-input-10-4dd6137cfe06>:3(create_array)
1 0.000 0.000 0.000 0.000 <ipython-input-10-4dd6137cfe06>:8(print_statement)
1 0.000 0.000 0.091 0.091 <string>:1(<module>)
3 0.000 0.000 0.000 0.000 iostream.py:195(schedule)
2 0.000 0.000 0.000 0.000 iostream.py:307(_is_master_process)
2 0.000 0.000 0.000 0.000 iostream.py:320(_schedule_flush)
2 0.000 0.000 0.000 0.000 iostream.py:382(write)
3 0.000 0.000 0.000 0.000 iostream.py:93(_event_pipe)
3 0.000 0.000 0.000 0.000 socket.py:357(send)
3 0.000 0.000 0.000 0.000 threading.py:1062(_wait_for_tstate_lock)
3 0.000 0.000 0.000 0.000 threading.py:1104(is_alive)
3 0.000 0.000 0.000 0.000 threading.py:506(is_set)
1 0.000 0.000 0.091 0.091 {built-in method builtins.exec}
2 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}
1 0.000 0.000 0.000 0.000 {built-in method builtins.print}
2 0.000 0.000 0.000 0.000 {built-in method posix.getpid}
3 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.lock' objects}
3 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}
400000 0.028 0.000 0.028 0.000 {method 'append' of 'list' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
Observe the above output. Notice that when a particular function is called more than once, the ncalls value reflects that. You can also spot the difference between the tottime and cumtime. This output clearly tells you that for i in range(0,400000) is the part where majority of time is spent.
5. How to use Profile class of cProfile
What is the need for Profile class when you can simply do a run()?
Even though the run() function of cProfile may be enough in some cases, there are certain other methods that are useful as well. The Profile() class of cProfile gives you more precise control. Let’s see a simple example.
By default, cProfile sorts its output by “standard name”. This means that it sorts by the filename(far right column). If you think of it, it’s actually not so useful, especially for complex functions. Also in case, the code contains a large number of steps, you cannot look through each line and find the time taken relatively.
How to use Profile to modify reports?
If your aim is to find the time-consuming parts, it would be helpful to sort the outputs as per ncalls. To do this,
- First, initialize an instance of Profile class.
- After that, call the
enable()method of the profiler to start collecting profiling data. - After that, call the function you want to profile.
- To stop collecting profiling data, call the
disable()method.
How to report the data collected ?
The pstats module can be used to manipulate the results collected by the profiler object. First, create an instance of the stats class using pstats.Stats. Next, use the Stats class to create a statistics object from a profile object through stats= pstats.Stats(profiler).Now, to sort the output by ncalls, use the sort_stats() method as shown below. Finally to print the output, call the function print_statss() of stats object.
# How to use Profile class of cProfile
def create_array():
arr=[]
for i in range(0,400000):
arr.append(i)
def print_statement():
print('Array created successfully')
def main():
create_array()
print_statement()
if __name__ == '__main__':
import cProfile, pstats
profiler = cProfile.Profile()
profiler.enable()
main()
profiler.disable()
stats = pstats.Stats(profiler).sort_stats('ncalls')
stats.print_stats()
Output:
Array created successfully
400039 function calls in 0.094 seconds
Ordered by: call count
ncalls tottime percall cumtime percall filename:lineno(function)
400000 0.034 0.000 0.034 0.000 {method 'append' of 'list' objects}
3 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.lock' objects}
3 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:93(_event_pipe)
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:195(schedule)
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/zmq/sugar/socket.py:357(send)
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1104(is_alive)
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:506(is_set)
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1062(_wait_for_tstate_lock)
2 0.000 0.000 0.000 0.000 {built-in method posix.getpid}
2 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:307(_is_master_process)
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:320(_schedule_flush)
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:382(write)
1 0.000 0.000 0.000 0.000 {built-in method builtins.print}
1 0.000 0.000 0.000 0.000 <ipython-input-1-66b56f7cc511>:6(print_statement)
1 0.004 0.004 0.094 0.094 <ipython-input-1-66b56f7cc511>:10(main)
1 0.055 0.055 0.090 0.090 <ipython-input-1-66b56f7cc511>:1(create_array)
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
You can see that the above output is different from previous and is sorted by ncalls. You can sort the output in various other ways.
Let’s say you want to sort the output by the cumulative time. You can do this by following the same code with a slight variation. Instead of ncalls, set sort_stats to use cumtime. The below code demonstrates it.
# Sort output by Cumulative time
if __name__ == '__main__':
import cProfile, pstats
profiler = cProfile.Profile()
profiler.enable()
main()
profiler.disable()
stats = pstats.Stats(profiler).sort_stats('cumtime')
stats.print_stats()
Output:
Array created successfully
400039 function calls in 0.088 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.004 0.004 0.088 0.088 <ipython-input-1-66b56f7cc511>:10(main)
1 0.057 0.057 0.083 0.083 <ipython-input-1-66b56f7cc511>:1(create_array)
400000 0.026 0.000 0.026 0.000 {method 'append' of 'list' objects}
1 0.000 0.000 0.000 0.000 <ipython-input-1-66b56f7cc511>:6(print_statement)
1 0.000 0.000 0.000 0.000 {built-in method builtins.print}
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:382(write)
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:195(schedule)
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/zmq/sugar/socket.py:357(send)
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1104(is_alive)
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:320(_schedule_flush)
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1062(_wait_for_tstate_lock)
2 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:307(_is_master_process)
3 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.lock' objects}
3 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:93(_event_pipe)
2 0.000 0.000 0.000 0.000 {built-in method posix.getpid}
3 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:506(is_set)
3 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}
2 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
Likewise, you can sort by other parameters such as percall, tottime and so on. Refer this https://docs.python.org/3.7/library/profile.html#pstats.Stats.sort_stats to know all options.
6. How to export cProfile data?
By default, the output of the profiler is simply printed out. But, you can use store the extracted data of profiling in a file as well. How to export the data/report?
The pstats module comes to use here.
After creating a Stats instance, pass the profiler as input to it as shown below. After that, use dump_stats() method to store it to any file by providing the path.
# Export profiler output to file
stats = pstats.Stats(profiler)
stats.dump_stats('/content/export-data')
Now, let’s consider a bit more lengthier example to organize the profiler output better. Let’s create a profile for the below code and print the report.
# Using cProfile.Profile example
import random
def print_msg():
for i in range(10):
print("Program completed")
def generate():
data = [random.randint(0, 99) for p in range(0, 1000)]
return data
def search_function(data):
for i in data:
if i in [100,200,300,400,500]:
print("success")
def main():
data=generate()
search_function(data)
print_msg()
if __name__ == '__main__':
import cProfile, pstats
profiler = cProfile.Profile()
profiler.enable()
main()
profiler.disable()
stats = pstats.Stats(profiler).sort_stats('tottime')
stats.print_stats()
Program completed
Program completed
Program completed
Program completed
Program completed
Program completed
Program completed
Program completed
Program completed
Program completed
5552 function calls in 0.003 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
1000 0.001 0.000 0.002 0.000 /usr/lib/python3.6/random.py:173(randrange)
1000 0.001 0.000 0.001 0.000 /usr/lib/python3.6/random.py:223(_randbelow)
1000 0.001 0.000 0.002 0.000 /usr/lib/python3.6/random.py:217(randint)
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:7(<listcomp>)
21 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/zmq/sugar/socket.py:357(send)
1268 0.000 0.000 0.000 0.000 {method 'getrandbits' of '_random.Random' objects}
1 0.000 0.000 0.000 0.000 <ipython-input-30-2a521dc30378>:10(search_function)
20 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:382(write)
1000 0.000 0.000 0.000 0.000 {method 'bit_length' of 'int' objects}
21 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:195(schedule)
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:15(main)
21 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1104(is_alive)
10 0.000 0.000 0.001 0.000 {built-in method builtins.print}
20 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:307(_is_master_process)
21 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.lock' objects}
21 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:1062(_wait_for_tstate_lock)
20 0.000 0.000 0.000 0.000 {built-in method posix.getpid}
1 0.000 0.000 0.001 0.001 <ipython-input-30-2a521dc30378>:3(print_msg)
21 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:93(_event_pipe)
20 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/ipykernel/iostream.py:320(_schedule_flush)
21 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}
20 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:6(generate)
21 0.000 0.000 0.000 0.000 /usr/lib/python3.6/threading.py:506(is_set)
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
If you observe, you might find the output cluttered and difficult to read. How can we improve this?
The pstats module provides the function strip_dirs() for this purpose. It removes all leading path information from file names.
# Remove dir names
stats.strip_dirs()
stats.print_stats()
Output:
5552 function calls in 0.003 seconds
Random listing order was used
ncalls tottime percall cumtime percall filename:lineno(function)
1000 0.000 0.000 0.000 0.000 {method 'bit_length' of 'int' objects}
20 0.000 0.000 0.000 0.000 {built-in method posix.getpid}
21 0.000 0.000 0.000 0.000 {method 'acquire' of '_thread.lock' objects}
1268 0.000 0.000 0.000 0.000 {method 'getrandbits' of '_random.Random' objects}
21 0.000 0.000 0.000 0.000 {method 'append' of 'collections.deque' objects}
20 0.000 0.000 0.000 0.000 {built-in method builtins.isinstance}
10 0.000 0.000 0.001 0.000 {built-in method builtins.print}
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:15(main)
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:6(generate)
1 0.000 0.000 0.000 0.000 <ipython-input-30-2a521dc30378>:10(search_function)
1 0.000 0.000 0.001 0.001 <ipython-input-30-2a521dc30378>:3(print_msg)
1 0.000 0.000 0.003 0.003 <ipython-input-30-2a521dc30378>:7(<listcomp>)
21 0.000 0.000 0.000 0.000 iostream.py:93(_event_pipe)
21 0.000 0.000 0.000 0.000 iostream.py:195(schedule)
20 0.000 0.000 0.000 0.000 iostream.py:307(_is_master_process)
20 0.000 0.000 0.000 0.000 iostream.py:320(_schedule_flush)
20 0.000 0.000 0.000 0.000 iostream.py:382(write)
21 0.000 0.000 0.000 0.000 socket.py:357(send)
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
1000 0.001 0.000 0.002 0.000 random.py:173(randrange)
1000 0.001 0.000 0.002 0.000 random.py:217(randint)
1000 0.001 0.000 0.001 0.000 random.py:223(_randbelow)
21 0.000 0.000 0.000 0.000 threading.py:1104(is_alive)
21 0.000 0.000 0.000 0.000 threading.py:506(is_set)
21 0.000 0.000 0.000 0.000 threading.py:1062(_wait_for_tstate_lock)
<pstats.Stats at 0x7f58db5659e8>
Observe the difference between the above and previous output. The above output is “random”. This is because, after a strip operation, the object has just been initialized and loaded.
7. How to visualize cProfile reports?
Even though we reduced some cluttering, there is still room to make it better. A good solution to get a clear picture of the profiling data is to visualize it.
A best tool available at the moment for visualizing data obtained by cProfile module is SnakeViz.
Let’s install it through the below command.
# Installing the module
!pip install snakeviz
Collecting snakeviz
[?25l Downloading https://files.pythonhosted.org/packages/a2/9a/6c753d20af6f177d3cbdb05a4b2e4419db4ec021c50ba86aa0d13a784a5c/snakeviz-2.1.0-py2.py3-none-any.whl (282kB)
[K |████████████████████████████████| 286kB 2.8MB/s
[?25hRequirement already satisfied: tornado>=2.0 in /usr/local/lib/python3.6/dist-packages (from snakeviz) (5.1.1)
Installing collected packages: snakeviz
Successfully installed snakeviz-2.1.0
For Ipython notebooks like google colab and Jupyter, you can load the SnakViz extension using %load_ext snakeviz command.
After this, call the function or program’s profiling you want to visualize through the %snakeviz <filename>. The filename can be either the entire python script or call to a particular function.
In the below code, I have written a main() function which calls several basic functions like creating an array and searching for specific elements. Now, to visualize the profiling data of the entire program I can use the command %snakeviz main() .
# Code to test visualization
import random
# Simple function to print messages
def print_msg():
for i in range(10):
print("Program completed")
# Generate random data
def generate():
data = [random.randint(0, 99) for p in range(0, 1000)]
return data
# Function to search
def search_function(data):
for i in data:
if i in [100,200,300,400,500]:
print("success")
def main():
data=generate()
search_function(data)
print_msg()
%load_ext snakeviz
%snakeviz main()
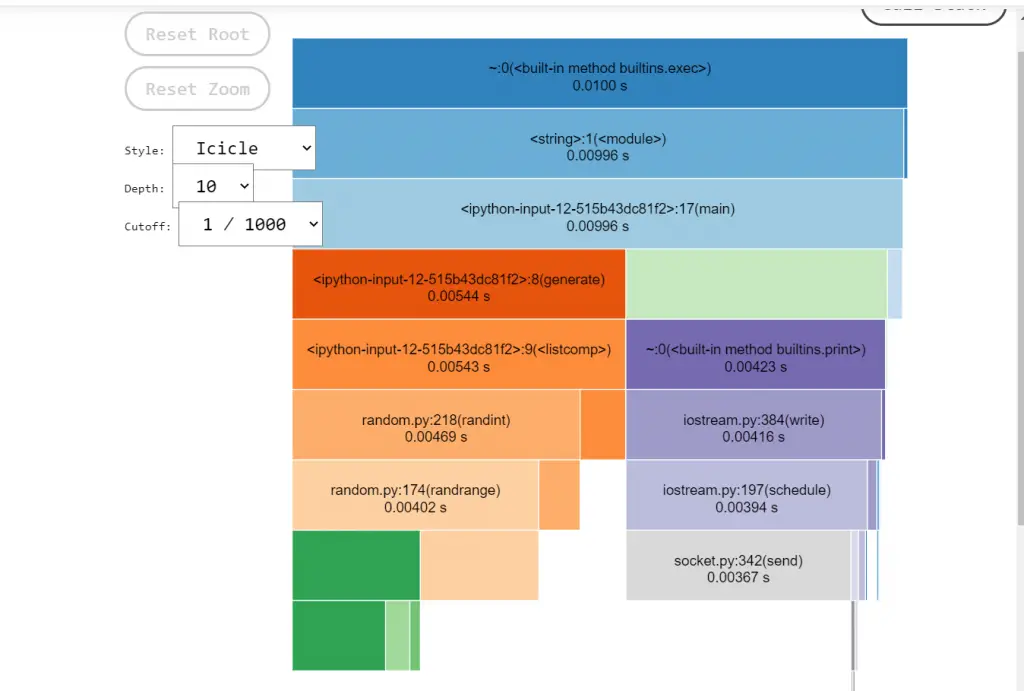
SnakeViz has two visualization styles, ‘icicle’ and ‘sunburst’.
By default, it’s icicle. icicle, the fraction of time taken by a code is represented by the width of the rectangle. Whereas in Sunburst, it is represented by the angular extent of an arc. You can switch between the two styles using the “Style” dropdown.
For the same code, let me show you the Sunburst style visualization too.
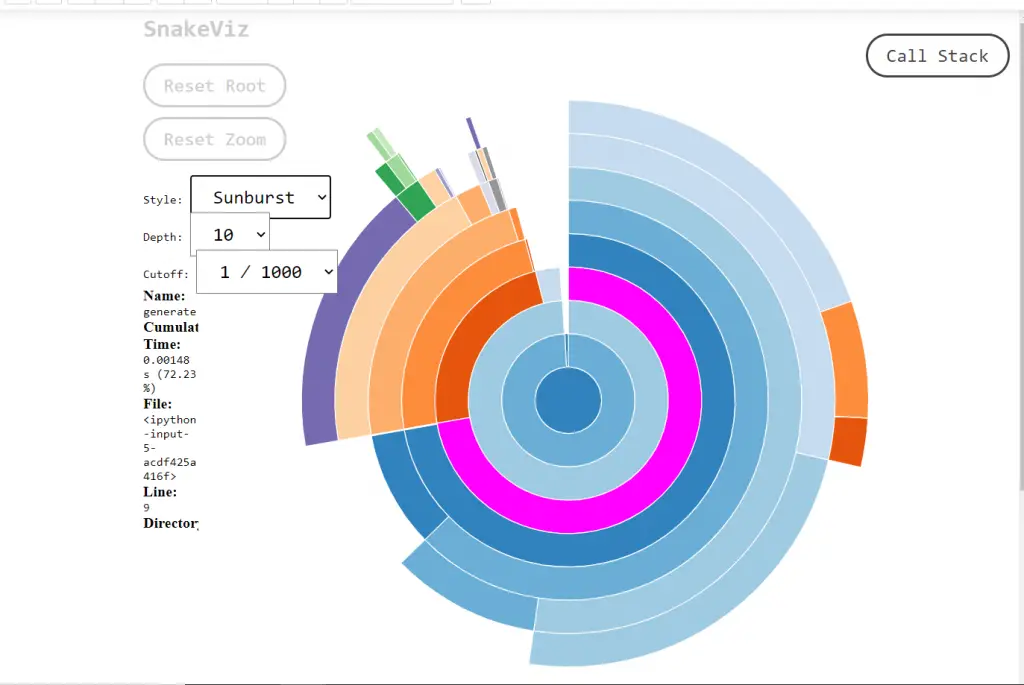
8. Profiling Linear Regression Model from scikit learn
Let’s look at a more complex example, where visualization would help a lot in real life.
Regression problems are very commonly used for various predictive modeling problems. The below code is a standard Linear regression problem using the sklearn library. Let’s print the profiling reports for this code.
# Function performing linear regression on diabetes dataset
def regression():
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import mean_squared_error, r2_score
# Load the diabetes dataset
diabetes_X, diabetes_y = datasets.load_diabetes(return_X_y=True)
# Use only one feature
diabetes_X = diabetes_X[:, np.newaxis, 2]
# Split the data into training/testing sets
diabetes_X_train = diabetes_X[:-20]
diabetes_X_test = diabetes_X[-20:]
# Split the targets into training/testing sets
diabetes_y_train = diabetes_y[:-20]
diabetes_y_test = diabetes_y[-20:]
# Create linear regression object
regr = linear_model.LinearRegression()
# Train the model using the training sets
regr.fit(diabetes_X_train, diabetes_y_train)
# Make predictions using the testing set
diabetes_y_pred = regr.predict(diabetes_X_test)
# Initialize profile class and call regression() function
profiler = cProfile.Profile()
profiler.enable()
regression()
profiler.disable()
stats = pstats.Stats(profiler).sort_stats('tottime')
# Print the stats report
stats.print_stats()
Output:
364724 function calls (357697 primitive calls) in 0.847 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
87 0.143 0.002 0.144 0.002 {built-in method _imp.create_dynamic}
379 0.110 0.000 0.110 0.000 {method 'read' of '_io.FileIO' objects}
739 0.088 0.000 0.089 0.000 /usr/lib/python3.6/inspect.py:2732(__init__)
379 0.055 0.000 0.055 0.000 {built-in method marshal.loads}
647/2 0.048 0.000 0.848 0.424 {built-in method builtins.exec}
1589 0.037 0.000 0.037 0.000 {built-in method posix.stat}
379 0.026 0.000 0.136 0.000 <frozen importlib._bootstrap_external>:830(get_data)
347 0.023 0.000 0.062 0.000 /usr/local/lib/python3.6/dist-packages/scipy/_lib/doccer.py:12(docformat)
809/806 0.019 0.000 0.031 0.000 {built-in method builtins.__build_class__}
1 0.013 0.013 0.013 0.013 /usr/local/lib/python3.6/dist-packages/scipy/linalg/lapack.py:784(_compute_lwork)
87/79 0.012 0.000 0.030 0.000 {built-in method _imp.exec_dynamic}
8222 0.010 0.000 0.010 0.000 {method 'splitlines' of 'str' objects}
561 0.010 0.000 0.012 0.000 <frozen importlib._bootstrap_external>:1080(_path_importer_cache)
375 0.010 0.000 0.016 0.000 /usr/local/lib/python3.6/dist-packages/scipy/_lib/doccer.py:179(indentcount_lines)
1 0.008 0.008 0.021 0.021 /usr/local/lib/python3.6/dist-packages/scipy/linalg/basic.py:1047(lstsq)
.................
6172 0.004 0.000 0.004 0.000 {method 'replace' of 'str' objects}
11729/11727 0.004 0.000 0.005 0.000 {method 'join' of 'str' objects}
391 0.004 0.000 0.095 0.000 /usr/lib/python3.6/inspect.py:2102(_signature_from_function)
4862 0.004 0.000 0.004 0.000 /usr/local/lib/python3.6/dist-packages/numpy/lib/npyio.py:790(floatconv)
101 0.003 0.000 0.173 0.002 /usr/local/lib/python3.6/dist-packages/scipy/stats/_distn_infrastructure.py:1572(__init__)
379 0.003 0.000 0.208 0.001 <frozen importlib._bootstrap_external>:743(get_code)
487/3 0.003 0.000 0.799 0.266 <frozen importlib._bootstrap>:966(_find_and_load)
36731/36652 0.003 0.000 0.003 0.000 {built-in method builtins.len}
554 0.003 0.000 0.003 0.000 {built-in method __new__ of type object at 0x9d12c0}
..................
466 0.002 0.000 0.010 0.000 <frozen importlib._bootstrap>:504(_init_module_attrs)
1563 0.002 0.000 0.002 0.000 {method 'format' of 'str' objects}
355 0.002 0.000 0.108 0.000 /usr/local/lib/python3.6/dist-packages/scipy/_lib/_util.py:277(getargspec_no_self)
2757 0.002 0.000 0.003 0.000 <frozen importlib._bootstrap_external>:59(<listcomp>)
2575/547 0.002 0.000 0.680 0.001 <frozen importlib._bootstrap>:997(_handle_fromlist)
36 0.002 0.000 0.002 0.000 {method 'read' of '_io.BufferedReader' objects}
1246 0.002 0.000 0.003 0.000 /usr/lib/python3.6/inspect.py:2452(__init__)
116 0.002 0.000 0.116 0.001 /usr/local/lib/python3.6/dist-packages/scipy/stats/_distn_infrastructure.py:622(_construct_argparser)
98 0.002 0.000 0.040 0.000 /usr/lib/python3.6/collections/__init__.py:357(namedtuple)
9839/9838 0.002 0.000 0.002 0.000 {built-in method builtins.isinstance}
105/31 0.002 0.000 0.005 0.000 /usr/lib/python3.6/sre_parse.py:470(_parse)
348 0.002 0.000 0.004 0.000 /usr/lib/python3.6/inspect.py:1787(_signature_bound_method)
580 0.002 0.000 0.004 0.000 /usr/lib/python3.6/functools.py:44(update_wrapper)
688 0.002 0.000 0.002 0.000 <frozen importlib._bootstrap>:103(release)
1 0.002 0.002 0.031 0.031 /usr/local/lib/python3.6/dist-packages/scipy/__init__.py:58(<module>)
.............
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/sparse/linalg/_expm_multiply.py:315(LazyOperatorNormInfo)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/sparse/linalg/_norm.py:3(<module>)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/optimize/_hessian_update_strategy.py:1(<module>)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/sparse/linalg/eigen/arpack/arpack.py:985(IterOpInv)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/sparse/linalg/interface.py:504(_CustomLinearOperator)
1 0.000 0.000 0.001 0.001 /usr/local/lib/python3.6/dist-packages/scipy/optimize/_trustregion_krylov.py:1(<module>)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/optimize/_trustregion_exact.py:188(IterativeSubproblem)
1 0.000 0.000 0.000 0.000 <string>:5(MetricInfo)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/special/sf_error.py:1(<module>)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/scipy/linalg/decomp_cholesky.py:1(<module>)
packages/joblib/externals/cloudpickle/cloudpickle.py:1139(_empty_cell_value)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/joblib/parallel.py:122(parallel_backend)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/joblib/parallel.py:322(BatchCompletionCallBack)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/joblib/_parallel_backends.py:578(SafeFunction)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-packages/joblib/my_exceptions.py:30(WorkerInterrupt)
1 0.000 0.000 0.000 0.000 /usr/local/lib/python3.6/dist-p
.....(truncated)...
<pstats.Stats at 0x7f58cf8bb2e8>
The above output has 207 entries!
In this case, visualization is not just an improvement option, but a definite necessity.
Let us apply SnakeViz visualization to the above code by calling the regression() function in the %snakeviz regression() command. This makes it very easier to interpret.
# Load the extension for visualizer.
%load_ext snakeviz
%snakeviz regression()
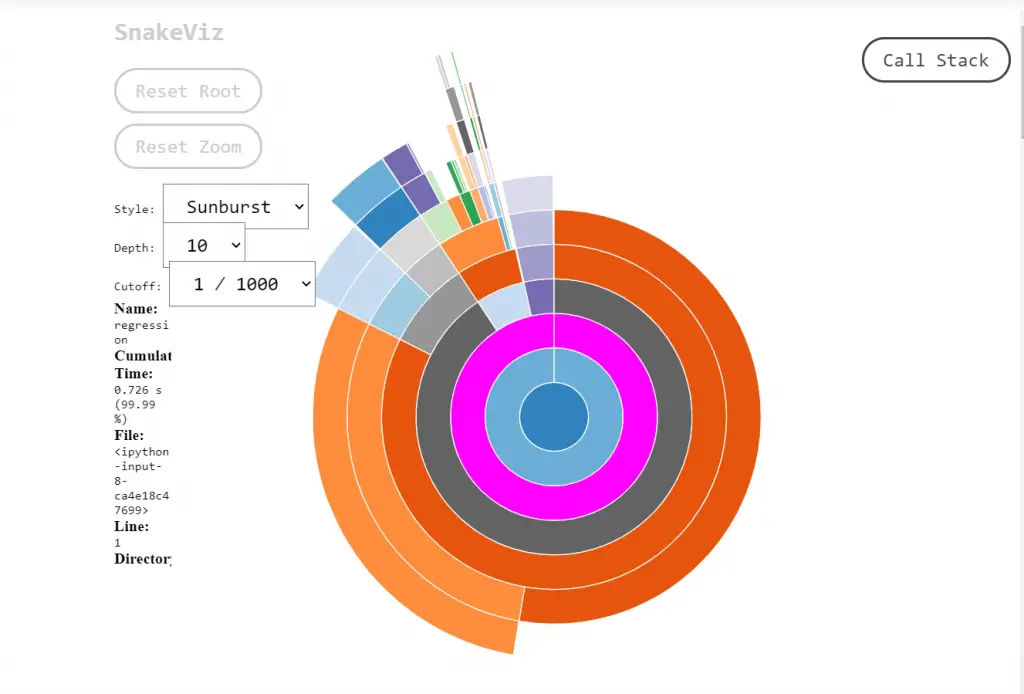
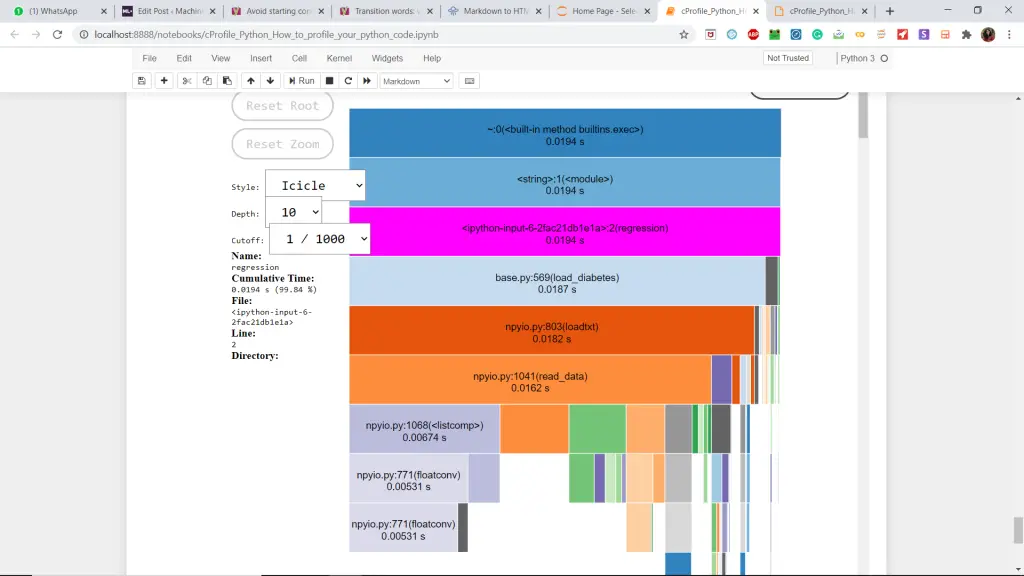
Note that you may not be able to get the visualizations properly in google colab. I recommend you to use Jupyter notebooks for convenience.
You can check other options available here for snakeviz.
9. Conclusion
I hope you understood the importance of profiling and how to use the cProfile module to get the statistics. Apart from cProfile, there is the Profile module of python too. But, cProfile includes most of its features and hence is recommended. Stay tuned for similar posts.






