data.table is a package is used for working with tabular data in R. It provides the efficient data.table object which is a much improved version of the default data.frame. It is super fast and has intuitive and terse syntax. If you know R language and haven’t picked up the `data.table` package yet, then this tutorial guide is a great place to start.
 Complete Beginners Guide to data.table in R. Photo by YaBoi.
Complete Beginners Guide to data.table in R. Photo by YaBoi.
Content
- Introduction: Why data.table?
- How to install data.table package
- Importing Data
- How to convert data.frame to data.table
- How to convert data.table to data.frame
- Filtering rows based on conditions
- How to select given columns
- How to select multiple columns using a character vector
- How to drop columns
- How to rename columns
- Creating a new column from existing columns
- How to create new columns using character vector
- Grouping
- What does .N and .I do
- Chaining
- What is .SD and How to write functions inside data.table
- Keys
- How to join two or more datatables
- How to merge multiple data.tables in one shot
- Pivot Table operations
- set() – A magic function for fast assignment operations
- Conclusion
1. Introduction: Why data.table?
The data.table is an alternative to R’s default data.frame to handle tabular data. The reason it’s so popular is because of the speed of execution on larger data and the terse syntax.
So, effectively you type less code and get much faster speed. It is one of the most downloaded packages in R and is preferred by Data Scientists. It is probably one of the best things that have happened to R programming language as far as speed is concerned.
Though data.table provides a slightly different syntax from the regular R data.frame, it is quite intuitive. So once you get it, it feels obvious and natural that you wouldn’t want to go back the base R data.frame syntax.
By the end of this guide you will understand the fundamental syntax of data.table and the structure behind it. All the core data manipulation functions of data.table, in what scenarios they are used and how to use it, with some advanced tricks and tips as well. data.table is authored by Matt Dowle with significant contributions from Arun Srinivasan and many others.
Related Post: 101 R data.table Exercises
2. How to install data.table package
Installing data.table package is no different from other R packages. Its recommended to run install.packages() to get the latest version on the CRAN repository. However, if you want to use the latest development version, you can get it from github as well.
# Install from CRAN
install.packages('data.table')
# Install Dev version from Gitlab
install.packages("data.table", repos="https://Rdatatable.gitlab.io/data.table")
data.table::update.dev.pkg()
If you want to revert back to the CRAN version do:
# Remove and Reinstall
remove.packages("data.table")
install.packages("data.table")
3. Importing Data
The way you work with data.tables is quite different from how you’d work with data.frames. Let’s understand these difference first while you gain mastery over this fantastic package.
The fread(), short for fast read is data.tables version of read.csv(). Like read.csv() it works for a file in your local computer as well as file hosted on the internet. Plus it is atleast 20 times faster. Let’s import the mtcars dataset stored as a csv file.
library(data.table)
mt <- fread("https://raw.githubusercontent.com/selva86/datasets/master/mtcars.csv")
head(mt)
class(mt)
#> [1] "data.table" "data.frame"
The imported data is stored directly as a data.table. As you see from the above output, the data.table inherits from a data.frame class and therefore is a data.frame by itself.
So, functions that accept a data.frame will work just fine on data.table as well. Because the dataset we imported was small, the read.csv()‘s speed was good enough.
However, the speed gain becomes evident when you import a large dataset (millions of rows). To get a flavor of how fast fread() is, run the below code. It creates a 1M rows csv file. Then reads it back again. The time taken by fread() and read.csv() functions gets printed in console.
# Create a large .csv file
set.seed(100)
m <- data.frame(matrix(runif(10000000), nrow=1000000))
write.csv(m, 'm2.csv', row.names = F)
# Time taken by read.csv to import
system.time({m_df <- read.csv('m2.csv')})
#> user system elapsed
#> 39.798 1.326 43.003
# Time taken by fread to import
system.time({m_dt <- fread('m2.csv')})
#> user system elapsed
#> 1.735 0.097 1.877
That’s about 20x faster. The time difference gets wider when the filesize increases.
4. How to convert data.frame to data.table
You can convert any `data.frame` into `data.table` using one of the approaches:
data.table(df)oras.data.table(df)setDT(df)
The difference between the two approaches is: data.table(df) function will create a copy of df and convert it to a data.table. Whereas, setDT(df) converts it to a data.table inplace.
That means, the df itself gets converted to a data.table and you don’t have to assign it to a different object.
As a result, there is no copy made and no duplication of the same data. Let’s reload the mtcars dataframe from R’s default datasets pacakge.
# reload data
data("mtcars")
head(mtcars)
Important: The data.table() does not have any rownames. So if the data.frame has any rownames, you need to store it as a separate column before converting to data.table.
mtcars$carname <- rownames(mtcars)
mtcars_dt <- as.data.table(mtcars)
class(mtcars_dt)
#> [1] "data.table" "data.frame"
Alternately, use setDT() to convert it to data.table in place.
mtcars_copy <- copy(mtcars)
setDT(mtcars_copy)
class(mtcars_copy)
#> [1] "data.table" "data.frame"
5. How to convert data.table to data.frame
Conversely, use as.data.frame(dt) or setDF(dt) to convert a data.table to a data.frame.
setDF(mtcars_copy)
class(mtcars_copy)
#> [1] "data.frame"
6. Filtering rows based on conditions
The main difference with data.frame is: data.table is aware of its column names. So while filtering, passing only the columns names inside the square brackets is sufficient.
# dataframe syntax
mtcars[mtcars$cyl == 6 & mtcars$gear == 4, ]
# datatable syntax
mtcars_dt[cyl==6 & gear==4, ]
This saves a significant amount of keystrokes in the long run. This is a major advantage. 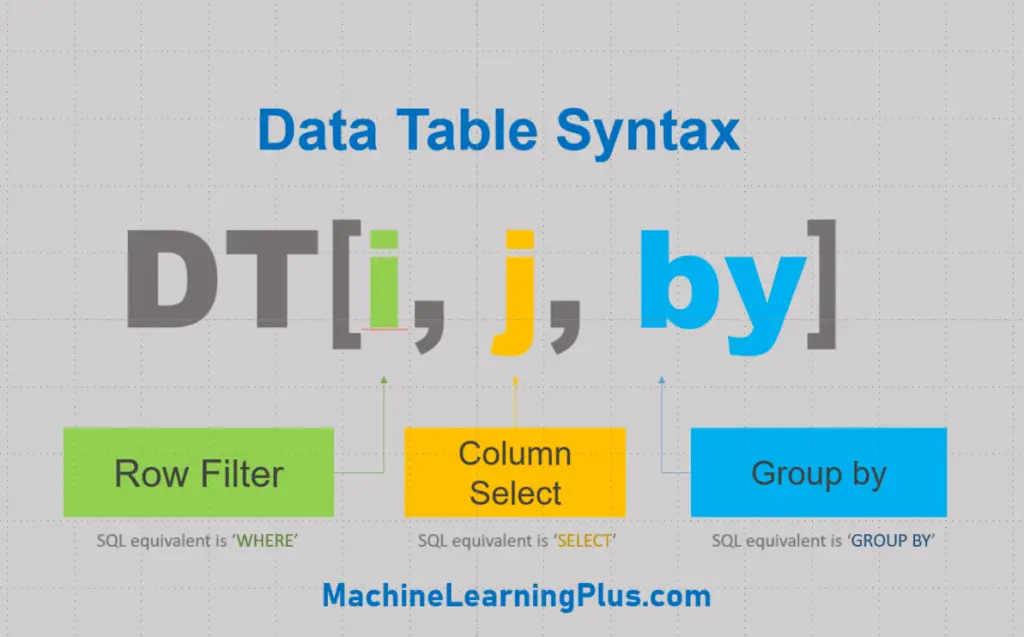
7. How to select given columns
Now, let see how to subset columns. The most unexpected thing you will notice with data.table is you cant select a column by its numbered position in a data.table. For example, you can expect the following to work in a data.frame.
mtcars[, 1]
# <returns first column>
But this would just return ‘1’ in a data.table
mtcars_dt[, 1]
#> 1
If you want to get that column by position alone, you should add an additional argument, with=FALSE.
mtcars_dt[, 1, with=F]
# <returns first column>
The returned output is a 1-column data.table. An alternate way and a better practice is to pass in the actual column name.
mtcars_dt[, mpg]
Notice here that the ‘mpg’ is not a string as it’s not written within quotes.
8. How to select multiple columns using a character vector
What if the column name is present as a string in another variable (vector)? In that case, you can’t use mpg directly. You need to additionally pass with=FALSE.
myvar <- "mpg"
mtcars_dt[, myvar, with=F]
# <returns mpg column>
The same principle applies if you have multiple columns to be selected.
columns <- c('mpg', 'cyl', 'disp')
mtcars_dt[, columns]
#> [1] "mpg" "cyl" "disp"
To make the above command work, you need to pass with=FALSE inside the square brackets
mtcars_dt[, columns, with=FALSE]
Clear? If you want to select multiple columns directly, then enclose all the required column names within list.
# syntax 1:
mtcars_dt[1:4, list(mpg, cyl, gear)]
# syntax 2: most used
mtcars_dt[, .(mpg, cyl, gear)]
9. How to drop columns
How to drop the mpg, cyl and gear columns alone? Place them in a vector and use the ! in front to drop them. This effectively returns all columns except those present in the vector.
drop_cols <- c("mpg", "cyl", "gear")
mtcars_dt[, !drop_cols, with=FALSE]
10. How to rename columns
The setnames() function is used for renaming columns. It takes the data.table (or data.frame), current name and new name as arguments and changes the column names in place without any copying of data. Always recommended!
setnames(mtcars_dt, 'vs', 'engine_type')
colnames(mtcars_dt)
#> <'vs' column is renamed to 'engine_type'.
DT <- data.table(A=1:5)
DT[ , X := shift(A, 1, type="lag")]
DT[ , Y := shift(A, 1, type="lead")]
Before moving on, let’s try out a mini challenge in R console. Q: Convert the in-built airquality dataset to a data.table. Then select “Solar.R”, “Wind” and “Temp” for those rows where “Ozone” is not missing. Show Solution
# Solution 1
aq_dt <- data.table(airquality)
aq_dt[!is.na(Ozone), .(Solar.R, Wind, Temp)]
# Solution 2
setDT(airquality)
airquality[!is.na(Ozone), .(Solar.R, Wind, Temp)]
11. Creating a new column from existing columns
You can always create a new column as you do with a data.frame, but, data.table lets you create column from within square brackets. This saves key strokes.
# data.frame syntax (works on data.table)
mtcars_dt$cyl_gear <- mtcars_dt$cyl + mtcars_dt$gear
# data.table syntax
mtcars_dt[, cyl_gear2 := cyl + gear]
mtcars_dt
To create multiple new columns at once, use the special assignment symbol as a function.
mtcars_dt[, `:=`(cyl_gear3 = cyl * gear,
cyl_gear4 = cyl - gear)]
mtcars_dt
To select only specific columns, use the list or dot symbol instead.
mtcars_dt[, .(cyl_gear3 = cyl * gear,
cyl_gear4 = cyl - gear)]
Now let’s see a special but frequently used case. Let’s suppose you have the column names in a character vector and want to select those columns alone from the data.table. Passing it inside the square brackets don’t work.
columns <- c('mpg', 'cyl', 'disp')
mtcars_dt[, columns]
#> [1] "mpg" "cyl" "disp"
To make the above command work, you need to pass with=FALSE inside the square brackets
mtcars_dt[1:3, columns, with=FALSE]
12. How to create new columns using character vector
Suppose you want to create a new column but you have the name of that new column in another character vector. How to create the new column without using the actual column name?
For example, you have the new column name in the myvar vector. And, you want to assign some value, say the value of 1 to this column. Doing this will create a new column named ‘myvar’. And not var1 as intended.
myvar <- c('var1')
mtcars_dt[, myvar:=1]
# <creates a column named myvar -- Bad! >
To create a column named ‘var1’ instead, keep myvar inside a vector.
# Create column named 'var1'
# Syntax 1
mtcars_dt[, c(myvar):=1]
# Syntax 2
mtcars_dt[, (myvar):=2]
Finally, if you want to delete a columns, assign it to NULL.
mtcars_dt[, c("myvar", "var1") := NULL]
All the above column names are now deleted. Before moving on, try solving this exercise in your R console. Question: Create a new column called ‘mileage_type’ that has the value ‘high’ if mpg > 20 else has value low. Show Solution
mtcars_dt[, mileage_type := ifelse(mpg > 20, 'high', 'low')]
13. Grouping
Now, let’s move on to the second major and awesome feature of R data.table: grouping using by. In base R, grouping is accomplished using the aggregate() function.
It’s a bit cumbersome and hard to remember the syntax.
All the functionalities can be accomplished easily using the ‘by’ argument within square brackets. For example, in mtcars data, how to get the mean mileage for each cylinder type? Answer: Since you want to see the mileage by cyl column, set by = 'cyl' inside the square brackets.
# Mean mileage by `cyl`
mtcars_dt[, .(mean_mileage=mean(mpg)), by=cyl]
Thats really useful isnt it? You can even add multiple columns to the ‘by’ argument.
mtcars_dt[, .(mean_mileage=mean(mpg)), by=.(cyl, gear)]
14. A slightly complex groupby problem
Now, lets see some really special cases.
How to select the first occurring value of mpg for each unique cyl value That is, instead of taking the mean of mileage for every cylinder, you want to select the first occurring value of mileage. How to do that?
mtcars_dt[, .(first_mileage=mpg[1]), by=cyl]
What to do if you want the second value? Just replace the 1 with 2.
mtcars_dt[, .(second_mileage=mpg[2]), by=cyl]
And what if you want the last value? You can either use length(mpg) or .N:
# Option 1
mtcars_dt[, .(first_mileage=mpg[length(mpg)]), by=cyl]
# Option 2
mtcars_dt[, .(first_mileage=mpg[.N]), by=cyl]
15. What does .N and .I do
.N contains the number of rows present. So the following will get the number of rows for each unique value of cyl.
mtcars_dt[, .N, by=cyl]
# < returns the number of rows for each unique value of cyl>
Now, how to create row numbers of items? It can be done using .I variable, short for ‘index’ (I guess). Lets first understand what .I returns.
mtcars_dt[, .I]
# <returns numbers from 1,2,3..>
It returns all the row numbers. Now, how to return the row numbers where cyl=6 ? This can get confusing in the beginning so pay close attention. If you want to get the row numbers of items that satisfy a given condition, you might tend to write like this:
mtcars_dt[cyl==6, .I]
# <again returns numbers from 1,2,3..>
But this returns the wrong answer because, `data.table` has already filtered the rows that contain cyl value of 6. So, what you want to do instead is to write that condition to subset .I alone instead of the whole `data.table`.
mtcars_dt[, .I[cyl==6]]
The result is same as using the `which()` function, which we used in `data.frames`.
mtcars_dt[, which(cyl==6)]
Great! Let’s solve another challenge before moving on. Q: Compute the number of cars and the mean mileage for each gear type. After a few seconds I will show the answer. Show Solution
mtcars_dt[, .(.N, mileage=mean(mpg) %>% round(2)), by=gear]
16. Chaining
Data.Table offers unique features there makes it even more powerful and truly a swiss army knife for data manipulation. First lets understand what chaining is.
Using chaining, you can do multiple datatable operatations one after the other without having to store intermediate results. For example, instead of writing two statements you can do it on one. Below code sorts after grouping by cyl:
dt1 <- mtcars_dt[, .(mean_mpg=mean(mpg),
mean_disp=mean(disp),
mean_wt=mean(wt),
mean_qsec=mean(qsec)), by=cyl]
output <- dt1[order(cyl), ]
output
With chaining, that is, by attaching the square brackets at the end, it’s done in one step.
output <- mtcars_dt[, .(mean_mpg=mean(mpg),
mean_disp=mean(disp),
mean_wt=mean(wt),
mean_qsec=mean(qsec)), by=cyl][order(cyl), ]
Actually, chaining is available in dataframes as well, but with features like by, it becomes convenient to use on a data.table.
17. What is .SD and How to write functions inside data.table
Next, lets see how to write functions within a data.table square brackets. Let’s suppose, you want to compute the mean of all the variables, grouped by ‘cyl’.
How to do that?
You can create the columns one by one by writing by hand. Or, you can use the lapply() function to do it all in one go. But `lapply()` takes the data.frame as the first argument. Then, how to use `lapply() inside a data.table? You can use the .SD object as the first argument for lapply(). But, what is the .SD object?
It is nothing but a data.table that contains all the columns of the original datatable except the column specified in ‘by’ argument. So, here is what it looks like.
mtcars_dt[, .SD, by=cyl]
# <returns the mtcars_dt table grouped by cyl. Try it>
So, now you can pass this as the first argument in `lapply()`. The 11th column in `.SD` is rownames, so let’s include only the first 10.
output <- mtcars_dt[, lapply(.SD[, 1:10, with=F], mean), by=cyl]
output
Optionally, Instead of subsetting .SD like this, You can specify the columns that should be part of .SD using the .SDCols object
output <- mtcars_dt[, lapply(.SD, mean), by=cyl, .SDcols=c("mpg", "disp", "hp", "drat", "wt", "qsec")]
output
The output now contains only the specified columns.
18. Keys
Now, we have come to the ‘key’ concept for data.tables: Keys
Let’s understand why keys can be useful and how to set it. Setting one or more keys on a data.table enables it to perform binary search, which is many order of magnitudes faster than linear search, especially for large data.
As a result, the filtering operations are super fast after setting the keys. There is a side effect though. By setting a key, the `data.table` gets sorted by that key. So how to set a key? Just use the setkey function.
setkey(mtcars_dt, carname)
It’s so fast making it look like nothing happened. But it internally sorted data.table with ‘carname’ as the key.
mtcars_dt
# <mtcars_dt is sorted by carname column which we created in beginning>
If you notice, this table is sorted by ‘carname’ variable. To check the keys for a data table, you can use the key() function.
key(mtcars_dt)
Once the key is set, merging data.tables is very direct. I have distributed few columns of mtcars in the following data.tables.
dt1 <- mtcars_dt[,.(carname, mpg, cyl)]
dt2 <- mtcars_dt[1:10, .(carname, gear)]
You can join these two datatables:
dt1[dt2]
This returns dt1‘s rows using dt2 based on the key of these data.tables. You can also set multiple keys if you wish.
setkey(mtcars_dt, cyl, mpg)
mtcars_dt
Now, how to remove the keys? Use setkey() and set it to NULL. But the datatable will not go back to it original row arrangement.
setkey(mtcars_dt, NULL)
Another aspect of setting keys is the ‘keyby’ argument. Using keyby you can do grouping and set the by column as a key in one go. For example, in this example we saw earlier, you can skip the chaining by using keyby instead of just by.
# Group and sort using chaining
output <- mtcars_dt[, .(mean_mpg=mean(mpg),
mean_disp=mean(disp),
mean_wt=mean(wt),
mean_qsec=mean(qsec)), by=cyl][order(cyl), ]
# Group and sort using keyby
output <- mtcars_dt[, .(mean_mpg=mean(mpg),
mean_disp=mean(disp),
mean_wt=mean(wt),
mean_qsec=mean(qsec)), keyby=cyl]
key(output)
As a result, the output has the key as cyl.
19. How to join two or more datatables
The data.table package provides a faster implementation of the merge() function. The syntax is pretty much the same as base R’s merge().
dt1 <- mtcars_dt[5:25,.(carname, mpg, cyl)]
dt2 <- mtcars_dt[1:10, .(carname, gear)]
dt3 <- mtcars_dt[2:12, .(carname, disp)]
# Inner Join
merge(dt1, dt2, by='carname')
#> <returns 6 rows>
# Left Join
merge(dt1, dt2, by='carname', all.x = T)
#> <returns 21 rows>
# Outer Join
merge(dt1, dt2, by='carname', all = T)
#> <returns 25 rows>
20. How to merge multiple data.tables in one shot
This is bit of a hack by using the Reduce() function to repeatedly merge multiple data.tables stored in a list. Reduce() takes in a function that has to be applied consequtively (which is merge_func in this case) and a list that stores the arguments for function.
# Merge multiple data.tables
dt_list <- list(dt1, dt2, dt3)
merge_func <- function(...) merge(..., all = TRUE, by='carname')
dt_merged <- Reduce(merge_func, dt_list)
#> <returns 25 rows from outer join>
21. Pivot Table operations
The dcast.data.table() is the function used for doing pivot table like operations as seen in spreadsheet softwares like Microsoft Office Excel or Google spreadsheets.
The good thing is dcast.data.table() works equally well on data.frame object as well. Let’s create a pivot table showing the mean mileage(mpg) for Cylinders vs Carburetter (Carb)
dcast.data.table(mtcars_dt, cyl ~ carb, fun.aggregate = mean, value.var = 'mpg')
So how to understand the syntax? There are 4 primary arguments:
- data.table
- formula: Rows of the pivot table on the left of ‘~’ and columns of the pivot on the right
- value.var: column whose values should be used to fill up the pivot table
- fun.aggregate: the function used to aggregate the
value.varcolumn.
dcast.data.table() is versatile in allowing multiple columns to be passed to the value.var and allows multiple functions to fun.aggregate as well. Let’s solve a quick exercise based on pivot table. Question 1: Create a pivot table to show the maximum and minimum mileage observed for Carburetters vs Cylinders combination? Question 2: Which carb value has the highest difference between max – min? Show Solution
# Solution for 1
dcast.data.table(mtcars_dt, carb ~ cyl, fun.aggregate = list(max, min), value.var = 'mpg')
# Solution for 2
dcast.data.table(mtcars_dt, carb ~ cyl, fun.aggregate = function(x)(max(x) - min(x)), value.var = 'mpg')
22. set() – A magic function for fast assignment operations
The set() command is an incredibly fast way to assign values to a new column. The syntax is: set(dt, i, j, value), where i is the row number and j is the column number.
As a best practice, always explicitly use integers for i and j, that is, use 10L instead of 10. It is usually used in for-loops and is literally thousands of times faster.
Yes, it is so fast even when used within a for-loop, which is proof that for-loop is not really a bottleneck for speed. It is the underlying data structure related overhead that causes for-loop to be slow, which is exactly what set() avoids. Besides, it works on a data.frame object as well.
It works like magic! Below is an example to illustrate the power of set() taken from official documentation itself. The speed benchmark may be outdated, but, run and check the speed by yourself to believe it.
m = matrix(1,nrow=100000,ncol=100)
DF = as.data.frame(m)
DT = as.data.table(m)
system.time(for (i in 1:10000) DF[i,1] <- i)
#> 591 seconds
system.time(for (i in 1:10000) DT[i,V1:=i])
#> 2.4 seconds ( 246 times faster, 2.4 is overhead in [.data.table )
system.time(for (i in 1:10000) set(DT,i,1L,i))
#> 0.03 seconds ( 19700 times faster, overhead of [.data.table is avoided )
22. Conclusion
We have covered all the core concepts in order to work with data.table package. To gain more practice, try the 101 R data.table Exercises. I’d be interested to know your comments as well, so please share your thoughts in the comments section below.






